
BITESIZEBIO CLC SEQUENCE VIEWER SOFTWARE
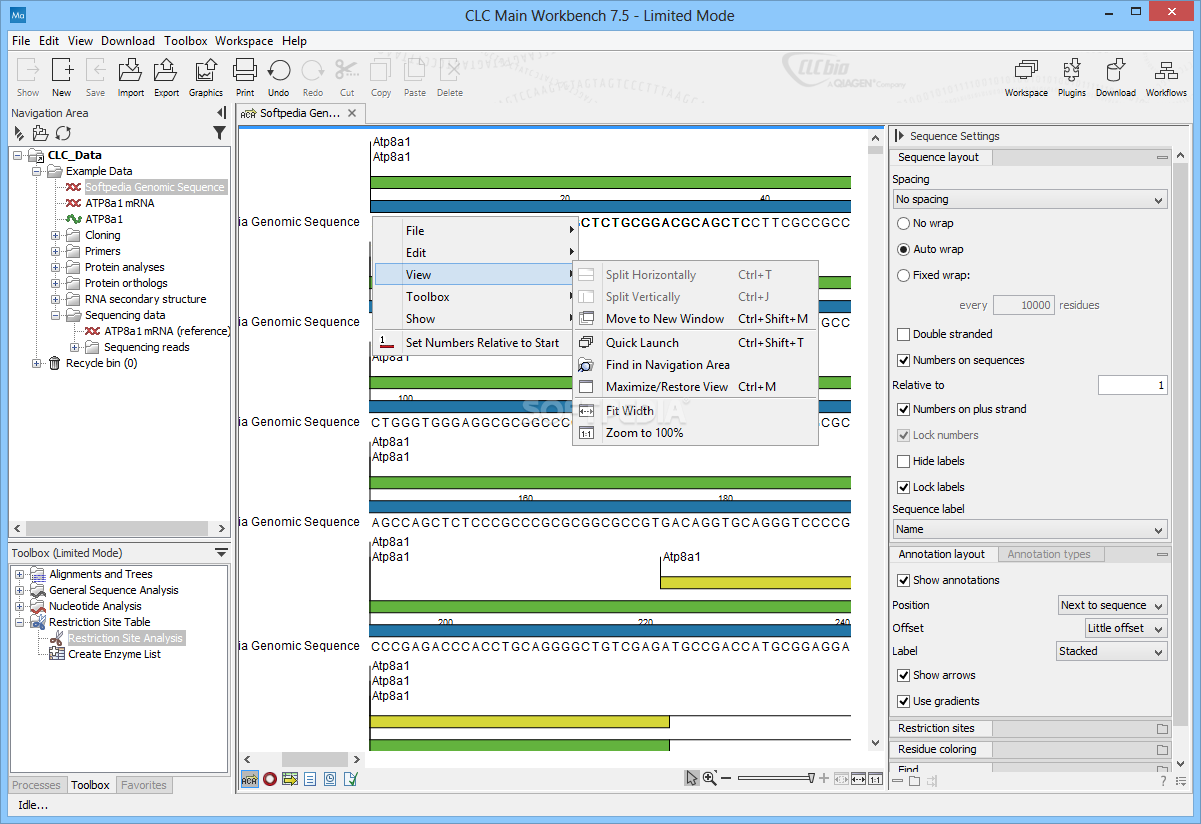
And what if the motif is on the opposite strand? Subsequently, your favorite sequence analysis software informs you that there is an interesting feature at position 13982–14013.
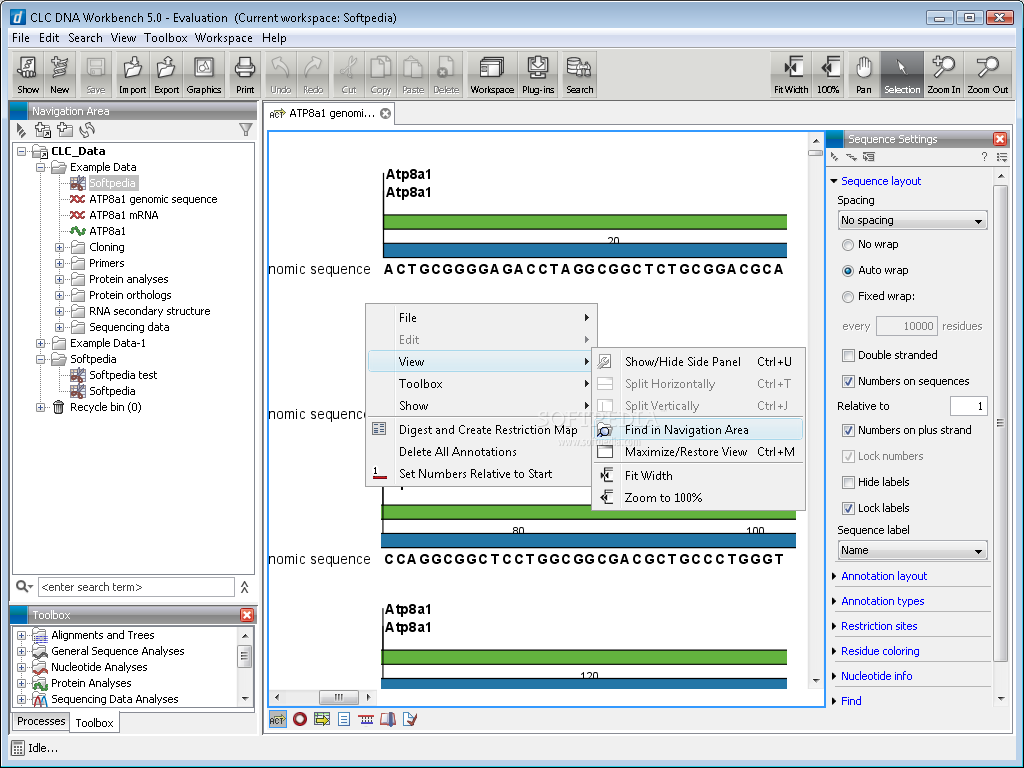
You reach for the browser's "find in page" button, but those darn spaces every 10 bp get in the way.

You are viewing a gene sequence in Entrez, and you want to find whether it contains a particular sequence motif. SeqVISTA: a graphical tool for sequence feature visualization and comparisonįull Text Available Abstract Background Many readers will sympathize with the following story. Our comparison study will assist researchers in selecting a well-suited assembler and offer essential information for the improvement of existing assemblers or the developing of novel assemblers. In terms of software implementation, string-based assemblers are superior to graph-based ones, of which SOAPdenovo is complex for the creation of configuration file. For large datasets of more than hundred millions of short reads, De Bruijn graph-based assemblers would be more appropriate. Considering the computational time, maximum random access memory (RAM occupancy, assembly accuracy and integrity, our study indicate that string-based assemblers, overlap-layout-consensus (OLC assemblers are well-suited for very short reads and longer reads of small genomes respectively. Here, we provide the information of adaptivity for each program, then above all, compare the performance of eight distinct tools against eight groups of simulated datasets from Solexa sequencing platform. Due to the poor knowledge about the applicability and performance of these software tools, choosing a befitting assembler becomes a tough task. The eShadow tool is publicly available at Ī practical comparison of de novo genome assembly software tools for next-generation sequencing technologies.ĭirectory of Open Access Journals (Sweden)įull Text Available The advent of next-generation sequencing technologies is accompanied with the development of many whole-genome sequence assembly methods and software, especially for de novo fragment assembly. Here, we describe the eShadow comparative tool and its potential uses for analyzing both multiple nucleotide and protein alignments to predict putative functional elements. This module grants the tool high flexibility in the analysis of multiple sequence alignments and in comparing sequences with different divergence rates. eShadow also includes a versatile optimization module capable of training the underlying Hidden Markov Model to differentially predict functional sequences. This tool integrates two different statistical methods and allows for the dynamic visualization of the resulting conservation profile. We have expanded this theoretical approach to create a computational tool, eShadow, for the identification of elements under selective pressure in multiple sequence alignments of closely related genomes, such as in comparisons of human to primate or mouse to rat DNA. Recently, a novel method, phylogenetic shadowing, has been pioneered for predicting functional elements in the human genome through the analysis of multiple primate sequence alignments. Primate sequence comparisons are difficult to interpret due to the high degree of sequence similarity shared between such closely related species. Ovcharenko, Ivan Boffelli, Dario Loots, Gabriela G. EShadow: A tool for comparing closely related sequencesĮnergy Technology Data Exchange (ETDEWEB)


 0 kommentar(er)
0 kommentar(er)
